January 12, 2021
National Institute of Infectious Diseases, JAPAN
Key Messages
- On January 6, 202, the National Institute of Infectious Diseases (NIID) of Japan detected a new variant isolate of SARS-CoV-2 from four travelers who arrived in Tokyo from Amazonas, Brazil, on January 2, 2021 at the airport screening. The isolate has some mutations found in previously reported variant isolates of concern from the UK and South Africa.
- Information on the variant isolate is limited to viral genome sequence data. Further investigation is necessary to assess infectivity, pathogenicity, and impact on laboratory diagnosis and vaccine efficacy of this variant strain.
- NIID recommends that persons infected with the variant isolate should be monitored in an isolated room and active epidemiological investigation should be initiated including contact tracing (with source investigation) and monitoring of the clinical course.
- Recommendations on prevention measures for individuals are avoidance of the three Cs, wearing a mask, and handwashing, as has been emphasized before.
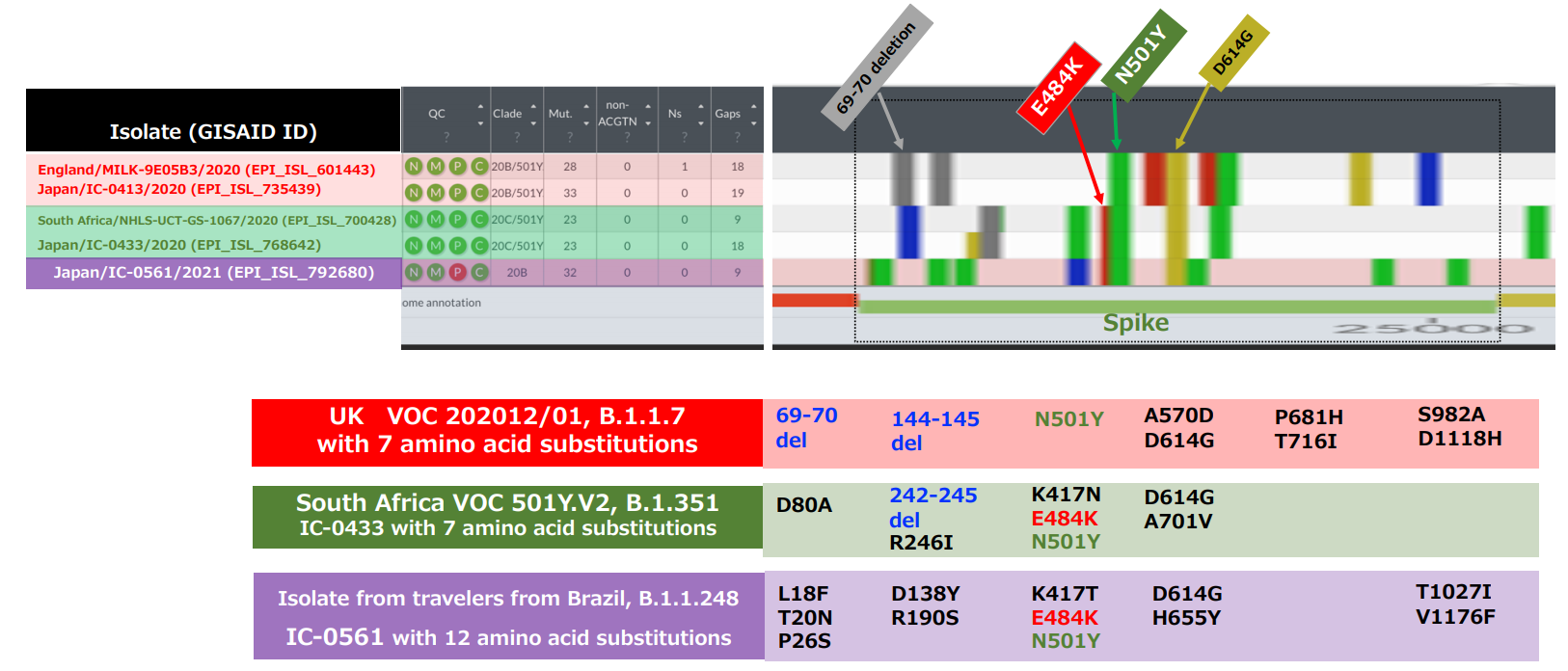
Technical detail
- The new variant isolate (GISAID ID: EPI_ISL_792680 to 792683) belongs to B.1.1.248 lineage and has 12 mutations in the spike protein, including N501Y and E484K.
- N501Y is a mutation found in variant strains including VOC-202012/01 and 501Y.V2, implicated to increase transmissibility.
- The E484K was reported to be an escape mutation from a monoclonal antibody which neutralize SARS-CoV-2 (1,2). The E484K has been observed in variant isolates escaping from convalescent plasma (3) and with a 10-fold decrease in neutralization capability by convalescent plasma (4) (both in preprint articles), suggesting possible change in antigenicity.
- In Brazil, a variant isolate with E484K belonging to B.1.1.248 was reported on January 6, 2021 (5), but it is not identical to the new variant isolate identified in Japan.
Reference
- Weisblum, Y. et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9, doi:10.7554/eLife.61312 (2020).
- Christian Gaebler, Zijun Wang, Julio C. C. Lorenzi, Frauke Muecksch, Shlomo Finkin, Minami Tokuyama, Mark Ladinsky, Alice Cho, Mila Jankovic, Dennis Schaefer-Babajew, Thiago Y. Oliveira, Melissa Cipolla, Charlotte Viant, Christopher O. Barnes, Arlene Hurley, Martina Turroja, Kristie Gordon, Katrina G. Millard, Victor Ramos, Fabian Schmidt, Yiska Weisblum, Divya Jha, Michael Tankelevich, Jim Yee, Irina Shimeliovich, Davide F. Robbiani, Zhen Zhao, Anna Gazumyan, Theodora Hatziioannou, Pamela J. Bjorkman, Saurabh Mehandru, Paul D. Bieniasz, Marina Caskey, Michel C. Nussenzweig. Evolution of Antibody Immunity to SARS-CoV-2. bioRxiv 2020.11.03.367391v2; doi: https://doi.org/10.1101/2020.11.03.367391
- Emanuele Andreano, Giulia Piccini, Danilo Licastro, Lorenzo Casalino, Nicole V. Johnson, Ida Paciello, Simeone Dal Monego, Elisa Pantano, Noemi Manganaro, Alessandro Manenti, Rachele Manna, Elisa Casa, Inesa Hyseni, Linda Benincasa, Emanuele Montomoli, Rommie E. Amaro, Jason S. McLellan, Rino Rappuoli. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020.12.28.424451; doi: https://doi.org/10.1101/2020.12.28.424451
- Allison J. Greaney, Andrea N. Loes, Katharine H.D. Crawford, Tyler N. Starr, Keara D. Malone, Helen Y. Chu, Jesse D. Bloom. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv 2020.12.31.425021; doi: https://doi.org/10.1101/2020.12.31.425021
- Vasques Nonaka, C.K.; Miranda Franco, M.; Gräf, T.; Almeida Mendes, A.V.; Santana de Aguiar, R.; Giovanetti, M.; Solano de Freitas Souza, B. Genomic Evidence of a Sars-Cov-2 Reinfection Case With E484K Spike Mutation in Brazil. Preprints 2021, 2021010132 (doi: 10.20944/preprints202101.0132.v1).